Possible
Jumping Genes In Pigeons
By Arif Mümtaz
February 2014
If you have been studying pigeon genetics, you should be familiar with
terms such as gene - a segment of
DNA that is responsible for the physical and
inheritable characteristics or
phenotype of an organism. DNA -
Deoxyribonucleic acid, a molecule that encodes the genetic instructions
used in
the development and function
of all known living organisms. Chromosome -
any of several threadlike linear strands of DNA, consisting of chromatin
that carry the genes in a linear order. DNA is made up of four similar
chemicals called bases A, T, C, and G, (Adenine, Thymine, Cytosine
Guanine) that are repeated over and over in pairs. Genes are located
on chromosomes and chromosomes
are passed down from generation to generation through
the egg and sperm. You should also be familiar
with the definition of mutation - changes
in the DNA sequence of a gene,
and chromosomal
crossover - the exchange of genetic material
between homologous chromosomes resulting in recombinant chromosomes.
Since
evolution is simply not possible without random genetic change for
its raw material, without mutation and crossover mechanisms, evolution
would have stopped dead.
 Our
knowledge of the genetic inheritance mechanism has grown immensely
after Darwin’s theory of evolution and Mendel’s laws
of inheritance. While the period from the early 1900s to World
War II has been considered the "golden age" of genetics,
scientists still had not determined that DNA, rather than protein,
is the hereditary
material. However, during this time there were a great many genetic
discoveries and the link between genetics and evolution was
made.
For the last two decades, we have entered what is known as the "genomic
era," where most of the scientific tests and discoveries
are made in
laboratory test tubes and sequencing of DNA is compared by
computer programs, instead of using traditional breeding data and Punnet
squares (classical
genetics) to obtain answers. The advances in molecular genetics
technology in the past two decades, particularly nucleic acid-based
markers, has had a great impact on gene mapping, allowing identification
of the underlying genes that control part of the variability of
traits. While breeding data for most of us is still important
and
will be used for many more years, research in molecular biology
and genetics has yielded answers to the basic questions left unanswered
by classical genetics about the make-up of genes, the mechanism
of
gene replication, what genes do, and the way that gene differences
bring about phenotypic differences. With the advent of DNA sequencing
technology and advanced studies in biochemistry of genes, we now
know there are many more exceptions to Mendelian genetics.
Our
knowledge of the genetic inheritance mechanism has grown immensely
after Darwin’s theory of evolution and Mendel’s laws
of inheritance. While the period from the early 1900s to World
War II has been considered the "golden age" of genetics,
scientists still had not determined that DNA, rather than protein,
is the hereditary
material. However, during this time there were a great many genetic
discoveries and the link between genetics and evolution was
made.
For the last two decades, we have entered what is known as the "genomic
era," where most of the scientific tests and discoveries
are made in
laboratory test tubes and sequencing of DNA is compared by
computer programs, instead of using traditional breeding data and Punnet
squares (classical
genetics) to obtain answers. The advances in molecular genetics
technology in the past two decades, particularly nucleic acid-based
markers, has had a great impact on gene mapping, allowing identification
of the underlying genes that control part of the variability of
traits. While breeding data for most of us is still important
and
will be used for many more years, research in molecular biology
and genetics has yielded answers to the basic questions left unanswered
by classical genetics about the make-up of genes, the mechanism
of
gene replication, what genes do, and the way that gene differences
bring about phenotypic differences. With the advent of DNA sequencing
technology and advanced studies in biochemistry of genes, we now
know there are many more exceptions to Mendelian genetics.
The history of inherited traits (gene concept) started with Gregor Mendel (1865-1910). Through his work with pea plants, Mendel discovered the fundamental laws of inheritance. In 1909 a Belgian Professor Frans Alfons Janssens of the University of Leuven described a phenomenon and called it "chiasmatypie". The term chiasma is thought to be what we now know as the chromosomal crossover. At that time, we knew genes were located on chromosomes, like beads on a string. Genes occupy certain "loci" on the chromosomes and are sometimes so closely linked together that they tend to "go together" in a common gamete. Thereby alleles at different loci that are closely linked sort in a dependent fashion, not necessarily independently, as Mendel assumed. Alleles of different genes ("loci") that are on different chromosomes or widely separated on the same chromosome sort independently, as Mendel assumed. In 1913 Thomas Hunt Morgan first described, in theory, crossing over of genes. When Morgan discovered the first Drosophila gene, he immediately saw the great importance of Janssens' cytological interpretation of chiasmata to the experimental results of his research on the heredity of Drosophila. Morgan was the founder of Drosophila genetics, and in his honor a recombination map unit is called a centiMorgan (cM). A map unit, or centiMorgan, is equal to crossing over between two genes in 1% of the gametes. Although crossing over of genes was first described by Morgan, the physical basis of crossing over was first demonstrated by Harriet Creighton and Barbara McClintock in 1931. McClintock produced the first genetic map for maize, linking regions of the chromosome to physical traits. She demonstrated the role of the telomere and centromere, regions of the chromosome that are important in the conservation of genetic information.
 In
addition to crossover of genes, in 1943, Dr. Barbara McClintock (pictured
on left) made an additional discovery that defied basic principles
of genetics;
this
discovery likely played a huge role in the course of evolution.
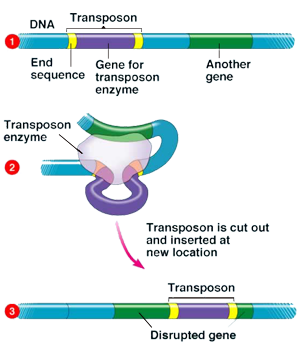
She discovered jumping genes (transposons)
which is a gene or set of genes capable of inserting copies of itself into
other DNA
sites
within the same cell. This discovery demonstrated that jumping
genes are responsible for turning physical characteristics on and off.
She
developed
theories
to explain the suppression and
expression of genetic information from one generation of maize
plants to the next. Although she was
a brilliant scientist, she faced many challenges; she was a member
of the Academy but was never offered an academic position
primarily because she was a woman. In
1942, disenchanted by her lack of career advancement in the male
dominated field of genetics, McClintock was offered a one-year
research position at the Carnegie Institution of Washington's
Department of Genetics in Cold Spring Harbor on Long Island, New York.
Working
alone, she observed differing coloration patterns in corn kernels
over generations of controlled studies.
In a series of
papers, she proposed that genes can travel on and
between chromosomes. Her theory of genetic transposition, commonly
called "jumping genes," explained that chromosomes
can break and recombine themselves. She saw that a break in the
chromosome
occurred when a gene randomly jumped or transposed from one chromosome
to another. When this happens, it disrupts the activity of
the other genes responsible for producing the pigment in the
kernel. Everybody at that time thought
the chromosomes and genes were very stable things just transmitted
from one generation to another.
McClintock
discovered that certain genes she was looking for were
in different positions in different corn plants. During the 1940s
and 1950s, skepticism of her research and its implications
developed and she stopped publishing her data in 1953. She eschewed
most scientific gatherings and had few extended conversations
or collaborations
with other scientists, a voluntary isolation which no doubt
slowed
the acceptance of her work. Her gender, of course, suggested
to many scientists that her work was of little consequence,
and that her
chosen area of expertise, maize (corn) was a further hindrance,
as it was of little interest to scientists of her time. She
was in no hurry to publish her findings in scientific journals,
and unveiled many years of her work in a single symposium at
Cold Spring Harbor in 1951. Even then, her work was generally
dismissed
until it was confirmed through use of improved molecular techniques
in the 1970s. Dr. McClintock died in 1992.
In
addition to crossover of genes, in 1943, Dr. Barbara McClintock (pictured
on left) made an additional discovery that defied basic principles
of genetics;
this
discovery likely played a huge role in the course of evolution.
She discovered jumping genes (transposons)
which is a gene or set of genes capable of inserting copies of itself into
other DNA
sites
within the same cell. This discovery demonstrated that jumping
genes are responsible for turning physical characteristics on and off.
She
developed
theories
to explain the suppression and
expression of genetic information from one generation of maize
plants to the next. Although she was
a brilliant scientist, she faced many challenges; she was a member
of the Academy but was never offered an academic position
primarily because she was a woman. In
1942, disenchanted by her lack of career advancement in the male
dominated field of genetics, McClintock was offered a one-year
research position at the Carnegie Institution of Washington's
Department of Genetics in Cold Spring Harbor on Long Island, New York.
Working
alone, she observed differing coloration patterns in corn kernels
over generations of controlled studies.
In a series of
papers, she proposed that genes can travel on and
between chromosomes. Her theory of genetic transposition, commonly
called "jumping genes," explained that chromosomes
can break and recombine themselves. She saw that a break in the
chromosome
occurred when a gene randomly jumped or transposed from one chromosome
to another. When this happens, it disrupts the activity of
the other genes responsible for producing the pigment in the
kernel. Everybody at that time thought
the chromosomes and genes were very stable things just transmitted
from one generation to another.
McClintock
discovered that certain genes she was looking for were
in different positions in different corn plants. During the 1940s
and 1950s, skepticism of her research and its implications
developed and she stopped publishing her data in 1953. She eschewed
most scientific gatherings and had few extended conversations
or collaborations
with other scientists, a voluntary isolation which no doubt
slowed
the acceptance of her work. Her gender, of course, suggested
to many scientists that her work was of little consequence,
and that her
chosen area of expertise, maize (corn) was a further hindrance,
as it was of little interest to scientists of her time. She
was in no hurry to publish her findings in scientific journals,
and unveiled many years of her work in a single symposium at
Cold Spring Harbor in 1951. Even then, her work was generally
dismissed
until it was confirmed through use of improved molecular techniques
in the 1970s. Dr. McClintock died in 1992.
The Nobel Prize for Physiology or Medicine was awarded to Dr. McClintock in 1983 for the discovery of genetic transposition. She is the only woman to receive an unshared Nobel Prize in that category. Since her discoveries, geneticists have been learning a lot more about DNA and how jumping genes affect the expression of genes. Until McClintock’s discoveries, genes were seen as inviolate objects with fixed positions (loci) on the chromosomes. We now know there are some genes (transposons) which do not have fixed positions in the genome and therefore can be removed from any cell; thus, all the progeny cells are back to their normal functions. In her observations of the chromosomal abnormalities, she noticed there were chromosome fragments left over from broken chromosomes. She figured out that what is known as the “ring” chromosomes were actually chromosomal fragments whose ends fused to form a ring structure. When she tracked the inheritance of these rings through mitosis, she found that they can be lost. McClintock concluded that different populations of corn cells can express different genes depending on when they keep or lose the ring chromosomes.
McClintock's study was a landmark in at least two directions: It showed that one gene can mutate another, and it illustrated that the genetic map with genes along a chromosome were capable of being changed by the action of another gene. McClintock noted unusual color patterns in the leaves of maize plants as they began to grow. She hypothesized that during cell division, certain cells lost genetic material, while others gained what they had lost. However, when comparing the chromosomes of the current generation of plants and their parent's generation, she found certain parts of the chromosomes had switched positions on the chromosome. McClintock's remarkable discovery, based entirely on her observation of chromosomes and genetic crosses at that time, had to wait for confirmation when we had the technology to sequence genomes.
 Although
you might be familiar with white or yellow corn, certain types of
corn naturally produce dark
purple
or blue
kernels. Tortillas
are made from the kernels of blue corn. McClintock’s
study of corn revealed that if a transposon moves to a position
neighboring
a pigment-producing
gene, the cells are unable to produce the normal purple pigment.
As a result of this, white streaked or mottled grain is produced,
rather than solid purple grain. If the pigmentation gene is turned
off long enough by a transposon, the grain will be completely unpigmented
(white colored). Therefore, the duration of a transposon in this "turned
off" position affects the degree of mottling. When a transposon
moves to different positions within cells of the corn kernel, the
coloration gene is "turned on" or "turned off" depending
on whether it lands in a position adjacent to the pigmentation gene.
This creates a variety of non-identical grain textures, which may
display blotches, dots, irregular lines and streaks. Thus,
the jumping genes are mobile genetic elements grouped under two classes
according to their mechanism
of transposition,
which
can be described
as either “copy and paste" for Class I TEs (retrotransposons)
or "cut and paste" for Class II TEs (transposons).
In other words, jumping genes (transposable elements) are
genes that move from
one location to another on a chromosome and change the
functionality of a working gene or completely make it none
functional by
turning the gene's function off.
Although
you might be familiar with white or yellow corn, certain types of
corn naturally produce dark
purple
or blue
kernels. Tortillas
are made from the kernels of blue corn. McClintock’s
study of corn revealed that if a transposon moves to a position
neighboring
a pigment-producing
gene, the cells are unable to produce the normal purple pigment.
As a result of this, white streaked or mottled grain is produced,
rather than solid purple grain. If the pigmentation gene is turned
off long enough by a transposon, the grain will be completely unpigmented
(white colored). Therefore, the duration of a transposon in this "turned
off" position affects the degree of mottling. When a transposon
moves to different positions within cells of the corn kernel, the
coloration gene is "turned on" or "turned off" depending
on whether it lands in a position adjacent to the pigmentation gene.
This creates a variety of non-identical grain textures, which may
display blotches, dots, irregular lines and streaks. Thus,
the jumping genes are mobile genetic elements grouped under two classes
according to their mechanism
of transposition,
which
can be described
as either “copy and paste" for Class I TEs (retrotransposons)
or "cut and paste" for Class II TEs (transposons).
In other words, jumping genes (transposable elements) are
genes that move from
one location to another on a chromosome and change the
functionality of a working gene or completely make it none
functional by
turning the gene's function off.
Class I and Class II TEs can be either autonomous or nonautonomous. Autonomous TEs can move on their own, while nonautonomous elements require other TEs presence in order to move. This is because nonautonomous elements lack the gene for the transposase or reverse transcriptase needed for their transposition, so they must "borrow" these proteins from another element in order to move (Pray, 2008).
 While
scientists are still discovering the effects of transposons on different
genes, we know transposons occasionally alter the activity of full-length
genes by turning on or off the neighboring genes. That activity occurs
more often in the brain than other areas, resulting in different traits
and behaviors, even in closely related individuals such as identical
twins.
Based on new discoveries, it turns out that transposons play a role
in people’s disposition to psychiatric disorders. Schizophrenia,
a pretty common mental disease is presumed to be controlled by the
transposons. Studies show that if one of the identical twins gets schizophrenia,
the other one have a 50% chance of getting it and a 50% chance of being
perfectly normal. Hence, if one twin happens to have one wild-type
and one schizophrenia type gene at that locus and a tronsposon turns
off
the wild-type copy, he/she becomes schizophrenic. If the transposon
turns off the schizophrenia type gene, that individual is normal. Which
copy
in heterozygotes that is turned off is random.
While
scientists are still discovering the effects of transposons on different
genes, we know transposons occasionally alter the activity of full-length
genes by turning on or off the neighboring genes. That activity occurs
more often in the brain than other areas, resulting in different traits
and behaviors, even in closely related individuals such as identical
twins.
Based on new discoveries, it turns out that transposons play a role
in people’s disposition to psychiatric disorders. Schizophrenia,
a pretty common mental disease is presumed to be controlled by the
transposons. Studies show that if one of the identical twins gets schizophrenia,
the other one have a 50% chance of getting it and a 50% chance of being
perfectly normal. Hence, if one twin happens to have one wild-type
and one schizophrenia type gene at that locus and a tronsposon turns
off
the wild-type copy, he/she becomes schizophrenic. If the transposon
turns off the schizophrenia type gene, that individual is normal. Which
copy
in heterozygotes that is turned off is random.
Today, scientists know that there are many different types of TEs, as well as a number of ways to categorize them. We now know that what McClintock had found in corn is also found in many plants and animals from algae to human beings. In fact, TEs make up approximately 50% of the human genome and up to 90% of the maize genome (SanMiguel, 1996). Although we are discovering that Transposon Elements are found in most life forms, we do not know how or when genes started to jump around the chromosomes. They may have originated from a common ancestor, or arisen independently multiple times, or arisen once and then spread to other kingdoms by horizontal gene transfer. What we do know is that while some TEs confer benefits to their host, most are regarded as selfish DNA parasites - similar to viruses. Because excessive TE activity can damage exons (active gene sequences), many organisms have developed mechanisms inhibiting their activities. Various viruses and TEs also share features in their genome structures and biochemical abilities leading to speculation that they share a common ancestor. Thus, it seems that over the course of evolution DNA transposons were deactivated, leaving them as introns (inactive gene sequences).
A mutation is a change in the genetic makeup of a living organism, and this change can either be beneficial or detrimental to the living organism. The main benefit of mutation is essentially survival; living organisms are currently alive due to successful mutations through natural selection. Crossover of genes occurs when two chromosomes, normally two homologous instances of the same chromosome, break and then reconnect but to the different end pieces. If they break at slightly different loci, the result can be a duplication of genes on one chromosome and a deletion of them on the other. Thus, crossover mechanism allows mutations to be linked on the same chromosome. Without the crossover, each gamete (sperm or egg) would get exact set of either paternal or maternal chromosomes. In other words, without crossover, all alleles for those genes linked together on the same chromosome would be inherited together. Thereby, the result of a crossover allows an equal amount of DNA material between pairs of chromosomes to be swapped from one chromosome to the other without changing overall sizes of each chromosome. The consequence of crossover is that the new chromosome may be superior to either of the parents if it takes the best characteristics (desirable traits) from each of the parents. It could also allow undesirable traits to be combined together. At any rate, without the mutation and crossover there cannot be evolution. But what role did transposons play over the course evolution?
For years jumping genes were basically ignored and considered “parasitic” or “junk” DNA. Now, they are being viewed as, perhaps, the most important mechanism of evolution. Plants appear to have the most jumping genes, with some genomes many times larger than humans with millions of self copied DNA elements. Corn has 40,000 genes, twice that of humans and 85% of their DNA is TEs. Since plants do not have as many behavioral responses as animals, it is possible they need more jumping genes to create evolutionary change in response to stress. At her Nobel Prize lecture, Dr. McClintock emphasized that transposition can provide a means to rapidly reorganize the genome in response to environmental stress. In this sense, mutations produced by transposition could be a source of variation driving the process of evolution. Transposons establish a hereditary relationship between two organisms from different species. Thus, by looking at the common transposon insertions, we can tell if different species share a common ancestry. The fact that transposable elements do not always excise perfectly and can take genomic sequences along for the ride has also resulted in a phenomenon scientists call exon shuffling. Exon shuffling results in the juxtaposition of two previously unrelated exons, usually by transposition, thereby potentially creating novel gene products (Moran et al., 1999). It seems we have barely scratched the surface of transposons, but one thing is certain: DNA transposons have the potential to influence the evolutionary trajectory of their hosts.
The genome is a pretty big sequence, so there are lots of places a transposon could reinsert. Sometimes a transposon will reinsert at another non-coding region leaving them as introns; this is usually what happens, since there’s so much more non-coding DNA than there is coding DNA. But sometimes a transposon can reinsert in a coding region and disrupt a gene which normally causes a debilitating mutation or disease. Consequently, we can argue that the transposon is as a separate selective entity to its host genome -- almost like a DNA parasite -- causing mostly non-beneficial mutations. Transposons also play a role in epigenetics by randomly turning the gene function on or off via alterations of gene function through insertion. However, there are also benefits which come from having transposons in a genome. Because transposon sequences occur randomly in different places in the genome, transposons offer an excellent way to identify individuals genetically. This becomes very relevant when DNA fingerprinting technology is used to establish paternity using a genetic test. These tests take advantage of the fact that two individuals in a population having the transposon sequences in the exact same location is extremely rare, so much so that we can conclude genetic relation based on similar patterns of genomic transposons.
 In
1999, cell biologists Dr. David Kirk, and Dr. Stephen Miller found
a transposon gene in in the green alga Volvox. Kirk and
Miller realized that by regulating the
temperature of its environment, they can make the Volvox gene jump
on command. This gene jumped so well and so
predictably when stimulated that Miller and Kirk
named it after their basketball hero Michael Jordan. They
found
that by growing cultures of Volvox at lower temperatures, they
could increase Jordan's jumping 30- to 50-fold. It is important to
have
Jordan jumping frequently because for every thousand times it jumps
in Volvox,
maybe only once will it land in or near a gene we're interested
in. There is no counterpart to a vertical leap in cell biology, but Kirk
and
Miller
found that Jordan jumps half the time to a nearby location and
the other half to random locations across the Volvox genome, or collection
of genes. There are five families of transposons found in Volvox,
but Jordan is the only one that can be made to jump. By studying
Jordan’s
jumps the scientists determined where it landed by tracking its
characteristic genetic signature. They used the jumping Jordan gene
to isolate genes
of interest and study their form and function. They recognized
the Jordan mutation in the gene where it landed, and then extracted
that
gene's preexisting DNA and studied it. The discovery of Jordan
on Volvox shed light on a simple organism that had been studied for
300 years,
and has implications for how organisms across species reproduce
and develop specialized cells.
In
1999, cell biologists Dr. David Kirk, and Dr. Stephen Miller found
a transposon gene in in the green alga Volvox. Kirk and
Miller realized that by regulating the
temperature of its environment, they can make the Volvox gene jump
on command. This gene jumped so well and so
predictably when stimulated that Miller and Kirk
named it after their basketball hero Michael Jordan. They
found
that by growing cultures of Volvox at lower temperatures, they
could increase Jordan's jumping 30- to 50-fold. It is important to
have
Jordan jumping frequently because for every thousand times it jumps
in Volvox,
maybe only once will it land in or near a gene we're interested
in. There is no counterpart to a vertical leap in cell biology, but Kirk
and
Miller
found that Jordan jumps half the time to a nearby location and
the other half to random locations across the Volvox genome, or collection
of genes. There are five families of transposons found in Volvox,
but Jordan is the only one that can be made to jump. By studying
Jordan’s
jumps the scientists determined where it landed by tracking its
characteristic genetic signature. They used the jumping Jordan gene
to isolate genes
of interest and study their form and function. They recognized
the Jordan mutation in the gene where it landed, and then extracted
that
gene's preexisting DNA and studied it. The discovery of Jordan
on Volvox shed light on a simple organism that had been studied for
300 years,
and has implications for how organisms across species reproduce
and develop specialized cells.
Since we are learning that all living organisms (both prokaryotes and eukaryotes) also have these transposable elements (TE's) in their DNA, we in the pigeon genetics community need to get familiar with this extraordinary phenomenon more popularly known as the “jumping genes” and add these words to our pigeon genetics lexicon. Attacking the genetic problems at the biochemical level after the Hollander Era has already started to pay off for pigeon genetics. With the start of the Genomic Era in pigeon genetics, results found in laboratory test tubes are already proving to be much more accurate than conventional Mendelian results. Biochemistry has allowed us to approach the genetic problems at the molecular level and, because the general plan of the macromolecules involved is rather simple, it has been remarkably successful. Nowadays, scientists look at both DNA sequencing data and biochemistry data, which are far more convincing than breeding data. Evidently, molecular genetics compliments the classical genetics (breeding data), but we are only at the beginning of discovering more accurate data from the sequencing of genes. Therefore, molecular geneticists at times find genotype data to be quite ugly as well. Yet, because of these exciting studies, we will soon be able to know which phenotypes are produced by the jumping genes when looking at the sequencing of a pigeon genome.
Although not widely accepted at the time of its discovery, Dr. McClintock's observation of the behavior of kernel color alleles was revolutionary in its proposition that genomic replication does not always follow a consistent pattern. Hence, at this point, it should be reasonable to guess which phenotypes in pigeons possibly result from transposons. My first reaction when I studied the jumping genes and saw the corn examples produced by this phenomenon was almond and grizzle phenotypes in pigeons. It is a pure guess on my part at this point to suggest that transposons may be the root cause of almond, grizzle, and their alleles. Although we do not yet have sequencing data available for neither of these genes, the idea certainly seems to be consistent with the observed phenotypes.
 |
 |
 |
 |
According to Dr. Smith, the transposable element may even code for the protein that does the moving. Further, it can have several copies of the transposable element in a row. Thus, it is not unreasonable to hypothesize that an allelic series like almond is caused by this phenomenon. The flecks causing various variegation of brown, black, and white in the almond series and the solid colored, or pure white, or mixed textured streaks in various grizzle phenotypes suggest a similarity to the transposons affect in corn. Although we have several hundred pigment genes in pigeons, it only takes one or two to be disrupted by a transposon to get almond or grizzle phenotypes. The other alleles of these mutants could easily be multiple copy issues of the original transposon or they could be something entirely different (private communication).
Although corn is indigenous to the western hemisphere, its exact birthplace is far less certain. Archeological evidence suggests that it was first found in Mexico. The original wild form has long been extinct but according to current studies, transposons make up 85% of the domesticated corn genome. Therefore, transposons may account for the rapid evolution of the tiny hard-kerneled teosinte form grown thousands of years ago in Mexico, to multi-colored Indian corn, and the larger and sweeter varieties popular today. Similar to dogs and domestic pigeons, corn has been rigorously selected over thousands of years and undergone incredible changes. Thus, it would not be perverse to theorize that transposable elements may also help to explain how the Rock pigeon genome can give rise to a variety of domestic pigeon breeds today. It might seem extreme and unjustified, but because of its performance variety (depth, velocity, frequency, style, and control), the rolling trait in Roller Pigeons might be due to transposons and/or other epigenetics. I for one, cannot wait to see the sequencing results of the roll gene(s). We certainly live in a very exciting time for new discoveries with the help of new technology. It is equally exciting to have scientists who have found pigeons to be once again the choice of species when experimenting with molecular genetics.
References:
1. Creighton H, McClintock B.
A Correlation of Cytological and Genetical Crossing-Over in Zea Mays.
Proc Natl Acad Sci USA 17 (8): 492–7 (1931).
2. Gage, Fred
H. & Muotri, Alysson R. Jumping Genes in the Brain Ensure
That Even Identical Twins Are Different. Scientific American, Volume
306. (2012).
3.
McClintock, B. Mutable loci in maize. Carnegie Institution of Washington
Yearbook 50, 174–181
(1951).
4. Miller, S. M. & Kirk, D. L. glsA, a Volvox gene required for asymmetric
division and germ cell specification, encodes a chaperone-like protein.
Development 126, 649–658 (1999).
5. Moran, J. V., et al. Exon shuffling
by L1 retrotransposition. Science 283, 1530–1534 (1999).
6. Pray, L. Transposons: The
jumping genes. Nature Education 1. (2008)
7.
SanMiguel, P., et al. Nested retrotransposons in the intergenic regions
of the maize genome. Science
274, 765–768 (1996).
8. Smith, Dan. PhD.
Private communication.
Copyright February, 2014 by Arif Mümtaz.